Coping with a few NP-Complete problems with Python
- Sandipan Dey

- Nov 17, 2020
- 11 min read
Updated: Nov 23, 2020
In this blog we shall discuss about a few NP-complete / NP-hard problems and some attempts to solve them (either by considering special cases and solving them efficiently, or by using algorithms to improve runtime or by using approximation algorithms and heuristics to obtain not necessarily optimal but good enough solutions) and the corresponding python implementations. The problems discussed here appeared as programming assignments in the coursera course Advanced Algorithms and Complexity and on Rosalind. The problem statements are taken from the course itself.
The Travelling Salesman Problem (TSP)
Improving the runtime of the Travelling Salesman Problem with Dynamic Programming
In this problem we shall deal with a classical NP-complete problem called Traveling Salesman Problem. Given a graph with weighted edges, you need to find the shortest cycle visiting each vertex exactly once. Vertices correspond to cities. Edges weights correspond to the cost (e.g., time) to get from one vertex to another one. Some vertices may not be connected by an edge in the general case.
Here we shall use dynamic programming to solve TSP: instead of solving one problem we will solve a collection of (overlapping) subproblems.
A subproblem refers to a partial solution
A reasonable partial solution in case of TSP is the initial part of a cycle
To continue building a cycle, we need to know the last vertex as well as the set of already visited vertices
It will be convenient to assume that vertices are integers from 1 to n and that the salesman starts his trip in (and also returns back to) vertex 1.
The following figure shows the Dynamic programming subproblems, the recurrence relation and the algorithm for TSP with DP.


Implementation tips
In order to iterate through all subsets of {1, . . . , n}, it will be helpful to notice that there is a natural one-to-one correspondence between integers in the range from 0 and 2^n − 1 and subsets of {0, . . . , n − 1}: k ↔ {i : i -th bit of k is 1}.
For example, k = 1 (binary 001) corresponds to the set {0}, where k = 5 (binary 101) corresponds to the set {0,2}
In order to find out the integer corresponding to S − {j} (for j ∈ S), we need to flip the j-th bit of k (from 1 to 0). For this, in turn, we can compute a bitwise XOR of k and 2^j (that has 1 only in j-th position)
In order to compute the optimal path along with the cost, we need to maintain back-pointers to store the path.
The following python code shows an implementation of the above algorithm.
import numpy as np
from itertools import combinations
def TSP(G):
n = len(G)
C = [[np.inf for _ in range(n)] for __ in range(1 << n)]
C[1][0] = 0 # {0} <-> 1
for size in range(1, n):
for S in combinations(range(1, n), size):
S = (0,) + S
k = sum([1 << i for i in S])
for i in S:
if i == 0: continue
for j in S:
if j == i: continue
cur_index = k ^ (1 << i)
C[k][i] = min(C[k][i], C[cur_index][j] + G[j][i])
#C[S−{i}][j]
all_index = (1 << n) - 1
return min([(C[all_index][i] + G[0][i], i) \
for i in range(n)])The following animation shows how the least cost solution cycle is computed with the DP for a graph with 4 vertices. Notice that in order to represent C(S,i) from the algorithm, the vertices that belong to the set S are colored with red circles, the vertex i where the path that traverses through all the nodes in S ends at is marked with a red double-circle.
The next animation also shows how the DP table gets updated. The DP table for a graph with 4 nodes will be of size 2^4 X 4, since there are 2^4=16 subsets of the vertex set V={0,1,2,3} and a path going through a subset of the vertices in V may end in any of the 4 vertex.
The transposed DP table is shown in the next animation, here the columns correspond to the subset of the vertices and rows correspond to the vertex the TSP ends at.

The following animation shows how the least cost solution cycle is computed with the DP for a graph with 5 nodes.

The following animation / figure shows the TSP optimal path is computed for increasing number of nodes (where the weights for the input graphs are randomly generated) and the exponential increase in the time taken.


Solving TSP with Integer Linear Program

Solving with the mip package using the following python code, produces the output shown by the following animation.
from mip import Model, xsum, minimize, BINARY
def TSP_ILP(G):
start = time()
V1 = range(len(G))
n, V = len(G), set(V1)
model = Model()
# binary variables indicating if arc (i,j) is used
# on the route or not
x = [[model.add_var(var_type=BINARY) for j in V] for i in V]
# continuous variable to prevent subtours: each city will have a
# different sequential id in the planned route except the first one
y = [model.add_var() for i in V]
# objective function: minimize the distance
model.objective = minimize(xsum(G[i][j]*x[i][j] \
for i in V for j in V))
# constraint : leave each city only once
for i in V:
model += xsum(x[i][j] for j in V - {i}) == 1
# constraint : enter each city only once
for i in V:
model += xsum(x[j][i] for j in V - {i}) == 1
# subtour elimination
for (i, j) in product(V - {0}, V - {0}):
if i != j:
model += y[i] - (n+1)*x[i][j] >= y[j]-n
# optimizing
model.optimize()
# checking if a solution was found
if model.num_solutions:
print('Total distance {}'.format(model.objective_value))
nc = 0 # cycle starts from vertex 0
cycle = [nc]
while True:
nc = [i for i in V if x[nc][i].x >= 0.99][0]
cycle.append(nc)
if nc == 0:
break
return (model.objective_value, cycle)
The constraint to prevent the subtours to appear in the solution is necessary, if we run without the constraint, we get a solution with subtours instead of a single cycle going through all the nodes, as shown below:


Comparing with Dynamic programming based solution, we can see that ILP is much more efficient for higher n values.


Bitonic TSP
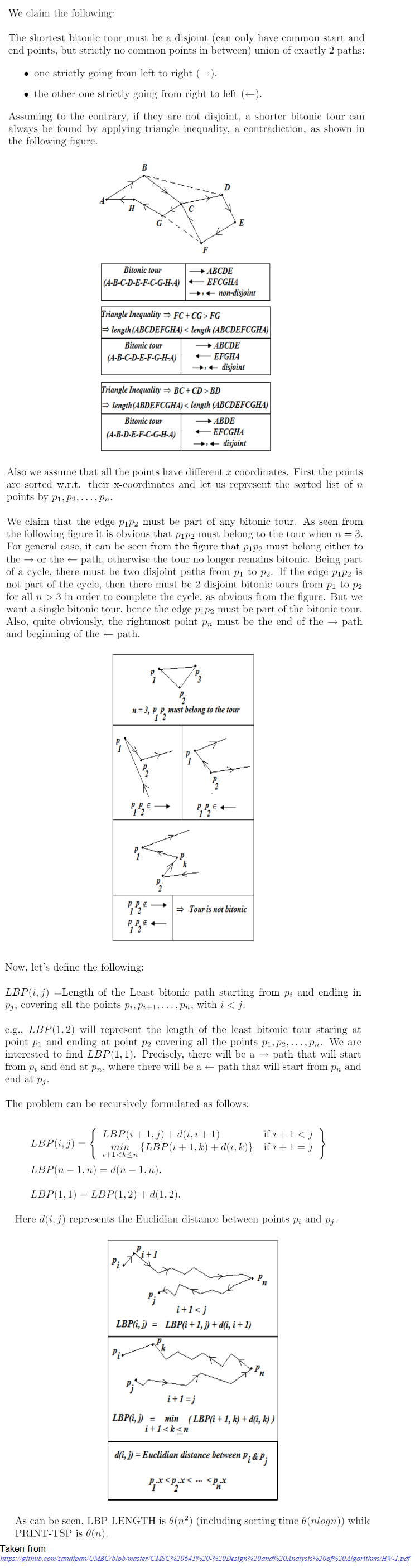

The following python code snippet implements the above DP algorithm.
def dist(P, i, j):
return np.sqrt((P[i][0]-P[j][0])**2+(P[i][1]-P[j][1])**2)
def BTSP(P):
n = len(P)
D = np.ones((n,n))*np.inf
path = np.ones((n,n), dtype=int)*(-1)
D[n-2,n-1] = dist(P, n-2, n-1)
path[n-2,n-1] = n-1
for i in range(n-3,-1,-1):
m = np.inf
for k in range(i+2,n):
if m > D[i+1,k] + dist(P,i,k):
m, mk = D[i+1,k] + dist(P,i,k), k
D[i,i+1] = m
path[i,i+1] = mk
for j in range(i+2,n):
D[i,j] = D[i+1,j] + dist(P,i,i+1)
path[i,j] = i+1
D[0,0] = D[0,1] + dist(P,0,1)
path[0,0] = 1
return D, path
def get_tsp_path(path, i, j, n):
if n < 0:
return []
if i <= j:
k = path[i,j]
return [k] + get_tsp_path(path, k, j, n-1)
else:
k = path[j,i]
return get_tsp_path(path, i, k, n-1) + [k]
The following animation shows how the DP table is computed and the optimal path for Bitonic TSP is constructed. It also shows the final optimal path.


2-OPT Approximation Algorithm for Metric TSP

The next code snippet implements the above 2-OPT approximation algorithm.
import numpy as np
import queue
def dfs(adj, x):
visited = [False]*len(adj)
stack = [x]
visited[x] = True
path = []
while len(stack) > 0:
u = stack.pop(-1)
path.append(u)
for v in adj[u]:
if not visited[v]:
stack.append(v)
visited[v] = True
return path
def mst(adj):
inf = np.inf
c = [inf]*n
s = 0
c[s] = 0
visited = [False]*n
parent = [None]*n
h = queue.PriorityQueue()
for v in range(n):
h.put((c[v], v))
edges = []
while not h.empty():
w, u = h.get()
if visited[u]: continue
visited[u] = True
if parent[u] != None:
edges.append((parent[u], u))
for v in range(n):
if v == u: continue
if (not visited[v]) and (c[v] > adj[u][v]):
c[v] = adj[u][v]
parent[v] = u
h.put((c[v], v))
adj = [[] for _ in range(n)]
for i in range(n):
if parent[i] != None:
adj[parent[i]].append(i)
path = dfs(adj, 0)
path += [path[0]]
return pathThe following animation shows the TSP path computed with the above approximation algorithm and compares with the OPT path computed using ILP for 20 points on 2D plane. The MST is computed with Prim's algorithm.

TSP with Simulated Annealing


The following python code snippet shows how to implement the Simulated Annealing to solve TSP, here G represents the adjacency matrix of the input graph.
def TSP_SA(G):
s = list(range(len(G)))
c = cost(G, s)
ntrial = 1
T = 30
alpha = 0.99
while ntrial <= 1000:
n = np.random.randint(0, len(G))
while True:
m = np.random.randint(0, len(G))
if n != m:
break
#print(ntrial, T, s)
s1 = swap(s, m, n)
c1 = cost(G, s1)
if c1 < c:
s, c = s1, c1
else:
if np.random.rand() < np.exp(-(c1 - c)/T):
s, c = s1, c1
T = alpha*T
ntrial += 1def swap(s, m, n):
i, j = min(m, n), max(m, n)
s1 = s.copy()
while i < j:
s1[i], s1[j] = s1[j], s1[i]
i += 1
j -= 1
return s1
def cost(G, s):
l = 0
for i in range(len(s)-1):
l += G[s[i]][s[i+1]]
l += G[s[len(s)-1]][s[0]]
return lThe following animations show how the algorithm works:


The following animation shows the TSP path computed with SA for 100 points in 2D.

TSP with Genetic Algorithm

Here in the following implementation of the above algorithm we shall have the following assumptions:
We shall assume the crossover rate is 1.0, i.e., all individuals in a population participate in crossover. The mutation probability to be used is 0.1.
With each crossover operation between two parent chromosomes, couple of children are generated, cant just swap portions of parents chromosomes, need to be careful to make sure that the offspring represents valid TSP path.
Mutation is similar to swap operation implemented earlier.
For each generation we shall keep a constant k=20 (or 30) chromosomes (representing candidate solutions for TSP).
The fitness function will be the cost of the TSP path represented by each chromosome. Hence, we want to minimize the value of the fitness function - i.e., less the value of a chromosome, more fit is it to survive.
We shall use rank selection, i.e., after crossover and mutation, only the top k fittest offspring (i.e., with least fitness function value) will survive for the next generation.
The following python code shows the implementation of the above algorithm with the above assumptions.
import numpy as np
def do_crossover(s1, s2, m):
s1, s2 = s1.copy(), s2.copy()
c1 = s2.copy()
for i in range(m, len(s1)): c1.remove(s1[i])
for i in range(m, len(s1)): c1.append(s1[i])
c2 = s1.copy()
for i in range(m, len(s2)): c2.remove(s2[i])
for i in range(m, len(s2)): c2.append(s2[i])
return (c1, c2)
def do_mutation(s, m, n):
i, j = min(m, n), max(m, n)
s1 = s.copy()
while i < j:
s1[i], s1[j] = s1[j], s1[i]
i += 1
j -= 1
return s1
def compute_fitness(G, s):
l = 0
for i in range(len(s)-1):
l += G[s[i]][s[i+1]]
l += G[s[len(s)-1]][s[0]]
return l
def get_elite(G, gen, k):
gen = sorted(gen, key=lambda s: compute_fitness(G, s))
return gen[:k]
def TSP_GA(G, k=20, ntrial = 200):
n_p = k
mutation_prob = 0.1
gen = []
path = list(range(len(G)))
while len(gen) < n_p:
path1 = path.copy()
np.random.shuffle(path1)
if not path1 in gen:
gen.append(path1)
for trial in range(ntrial):
gen = get_elite(G, gen, k)
gen_costs = [(round(compute_fitness(G, s),3), s) for s in gen]
next_gen = []
for i in range(len(gen)):
for j in range(i+1, len(gen)):
c1, c2 = do_crossover(gen[i], gen[j], \
np.random.randint(0, len(gen[i])))
next_gen.append(c1)
next_gen.append(c2)
if np.random.rand() < mutation_prob:
m = np.random.randint(0, len(gen[i]))
while True:
n = np.random.randint(0, len(gen[i]))
if m != n:
break
c = do_mutation(gen[i], m, n)
next_gen.append(c)
gen = next_gen
The following animation shows the TSP path computed with GA for 100 points in 2D.

Coloring a Graph with 3 colors using a SAT solver
Given a graph, we need to color its vertices into 3 different colors, so that any two vertices connected by an edge need to be of different colors. Graph coloring is an NP-complete problem, so we don’t currently know an efficient solution to it, and you need to reduce it to an instance of SAT problem which, although it is NP-complete, can often be solved efficiently in practice using special programs called SAT-solvers.
We can reduce the real-world problem about assigning frequencies to the transmitting towers of the cells in a GSM network to a problem of proper coloring a graph into 3 colors.Colors correspond to frequencies, vertices correspond to cells, and edges connect neighboring cells, as shown below.


We need to output a boolean formula in the conjunctive normal form (CNF) in a specific format. If it is possible to color the vertices of the input graph in 3 colors such that any two vertices connected by an edge are of different colors, the formula must be satisfiable. Otherwise, the formula must be unsatisfiable.
We shall use pySAT SAT-solver to solve the clauses generated from the graph.
In particular, if there are n nodes and m edges in the graph that is to be colored with 3 colors, we need to generate 3*n variables and 3*m + n clauses.
The i-th node will correspond to 3 variables, with id i, n+i and 2*n+i. They will represent whether the node is to be colored by red, green or blue.
Any two vertices forming an edge must not have the same color.
Each node must be colored by one from the 3 colors.
The following python code snippet shows how the encoding needs to be implemented to output the desired clauses for the CNF, subsequently solved by the SAT-solver and the solution assignments is used to color the vertices of the graph. Again, there are following 3 basic steps:
encode the input graph into SAT formula (reduce the 3-coloring to a satisfiability problem)
solve with a SAT and obtain solution in terms of variable assignments
use the solution assignment by SAT to color the graph
import numpy as np
from pysat.solvers import Glucose3
def get_colors(assignments):
all_colors = np.array(['red', 'green', 'blue'])
colors = {}
for v in range(n):
colors[v+1] = all_colors[[assignments[v]>0, \
assignments[n+v]>0, assignments[2*n+v]>0]][0]
return colors
def print_clauses(clauses):
for c in clauses:
vars = []
for v in c:
vars.append('{}x{}'.format('¬' if v < 0 else '', abs(v)))
print('(' + ' OR '.join(vars) + ')')
def print_SAT_solution(assignments):
sol = ''
for x in assignments:
sol += 'x{}={} '.format(abs(x),x>0)
print(sol)
def solve_graph_coloring():
# encode the input graph into SAT formula
n_clauses = 3*m+n
n_vars = 3*n
clauses = []
for u, v in edges:
clauses.append((-u, -v)) # corresponding to red color
clauses.append((-(n+u), -(n+v))) # corresponding to green color
clauses.append((-(2*n+u), -(2*n+v))) # corresponds to blue colr
for v in range(1, n+1):
clauses.append((v, n+v, 2*n+v)) # at least one color
print_clauses(clauses)
# solve SAT and obtain solution in terms of variable assignments
g = Glucose3()
for c in clauses:
g.add_clause(c)
status = g.solve()
assignments = g.get_model()
print(status)
print_SAT_solution(assignments)
# use the solution assignment by SAT to color the graph
colors = get_colors(assignments)
print(colors)The following animations show the input graphs, the corresponding variables and clauses generated for the CNF to be solved with the SAT-solver, the solution obtained in terms of truth-assignments of the variables and then how they are used solve the graph-coloring problem, to color the graph with 3 colors. Each row in the right subgraph represents a clause in the CNF.


The last graph is the Petersen graph, that has chromatic number 3, so can be colored with 3 colors and a 3-coloring solution is obtained by the SAT-solver output assignments.

Now, the following example shows an input graph which is not 3-colorable, with the CNF formed by the clauses generated to be solved by the SAT-solver being unsatisfiable / inconsistent.

Solving the Hamiltonian Path problem with SAT-solver
In this problem, we shall learn how to solve the classic Hamiltonian Path problem, by designing and implementing an efficient algorithm to reduce it to SAT.

The following python snippet shows:
1. how the Hamiltonian Path problem is reduced to SAT
2. Then it's solved by the pysat SAT-solver.
3. The solution is interpreted to construct the Hamiltonian path for the following input graphs.
def get_hamiltonian_path(assignments):
path = [None]*n
for i in range(n):
for j in range(n):
if assignments[i*n+j] > 0: # True
path[i] = j+1
return path
def reduce_Hamiltonian_Path_to_SAT_and_solve(edges):
def index(i, j):
return n*i + j + 1
m = len(edges)
n_clauses = 2*n + (2*n*n-n-m)*(n-1)
n_vars = n*n
clauses = []
for j in range(n):
clause = []
for i in range(n):
clause.append(index(i,j))
clauses.append(clause)
for i in range(n):
clause = []
for j in range(n):
clause.append(index(i,j))
clauses.append(clause)
for j in range(n):
for i in range(n):
for k in range(i+1, n):
clauses.append((-index(i,j), -index(k,j)))
for i in range(n):
for j in range(n):
for k in range(j+1, n):
clauses.append((-index(i,j), -index(i,k)))
for k in range(n-1):
for i in range(n):
for j in range(n):
if i == j: continue
if not [i+1, j+1] in edges:
clauses.append((-index(k,i), -index(k+1,j)))
print_clauses(clauses)
g = Glucose3()
for c in clauses:
g.add_clause(c)
status = g.solve()
assignments = g.get_model()
print(status)
print_SAT_solution(assignments)
path = get_hamiltonian_path(assignments)
print(path)The following figure shows the output Hamiltonian Path obtained for the line input graph using the solution obtained by SAT-solver.

The following figure shows the Hamiltonian Path obtained with the SAT-solver for the Petersen's graph, which indeed has a Hamiltonian Path.






Comments